Research
Our laboratory is working on the following major research topics to understand and apply the defense mechanisms against viral infections.
Human innate immune response to viral infection
Our body is equipped with a sophisticated immune mechanism to recognize pathogens such as viruses and bacteria as foreign substances and eliminate them. Immunity is largely divided into innate immunity and acquired immunity. In particular, innate immunity does not recognize host-derived “self” molecules via receptor proteins, but specifically recognizes characteristics of pathogen molecules as “non-self. In our laboratory, our research focuses on immune receptors that exist within cells.
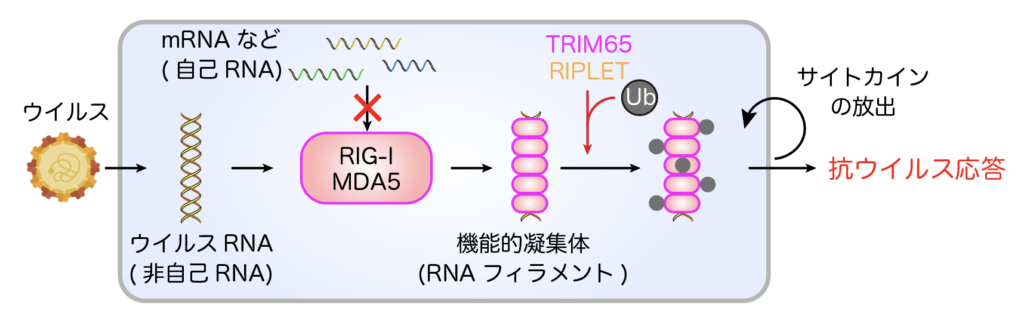
RIG-I-like receptors (RIG-I, MDA5) recognize viral RNA that has entered the cell and form aggregate structures called RNA filaments (Figure 1). This activates downstream signaling and the production of potent cytokines called interferons, and the host acquires an antiviral state. This distinction between “self” and “non-self” must be strictly controlled, and its breakdown leads to the development of autoimmune diseases.
We are currently working on the following projects.
1. Elucidation of the mechanism by which RIG-I and MDA5 strictly distinguish self/nonself RNAs and form aggregate structures
It has become clear that immune responses by RIG-I and MDA5 involve various cofactor proteins and post-translational modifications, and that immune responses occur only against “non-self” viral RNA (Kato et al., Mol Cell, 2021, https://www.cell. com/molecular-cell/pdf/S1097-2765(20)30889-3.pdf). In other words, immune responses by RIG-I and MDA5 are hierarchically regulated by multiple checkpoints (Figures 2 and 3). Our research focuses on the mechanisms by which these cofactor proteins regulate RIG-I/MDA5 function
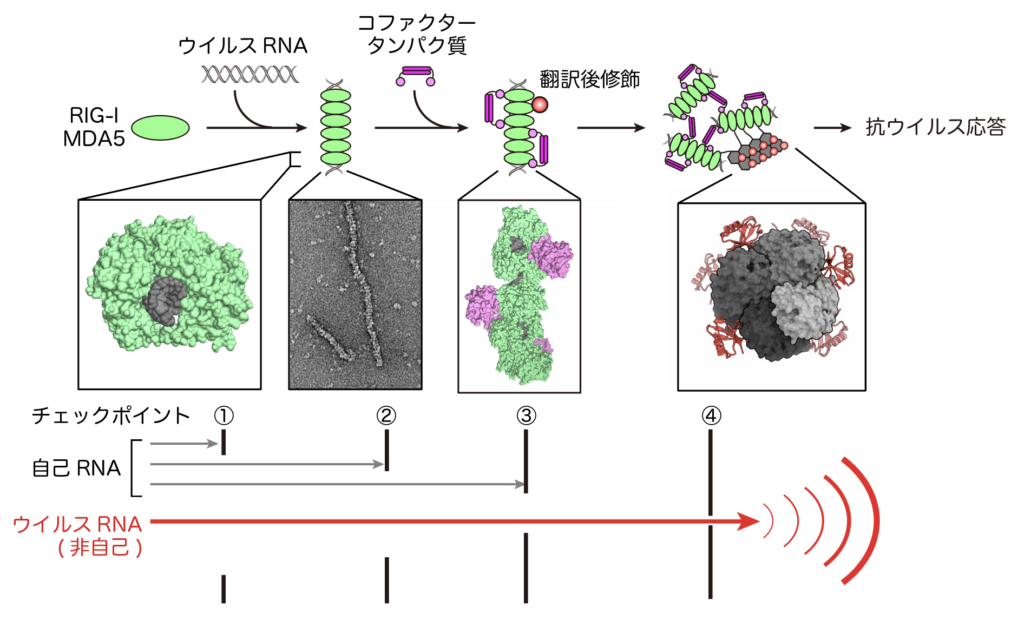
In the antiviral immune response by RIG-I/MDA5, there are multiple checkpoints to distinguish between “self” and “non-self” RNA. (i) Specific recognition of structures characteristic of viral RNA. (ii) It recognizes the length of viral RNA by forming aggregates (filaments). Cofactor proteins (TRIM65 and RIPLET) recognize only RNA filaments and perform post-translational modification (ubiquitination). (iv) Ubiquitination enhances downstream signaling.
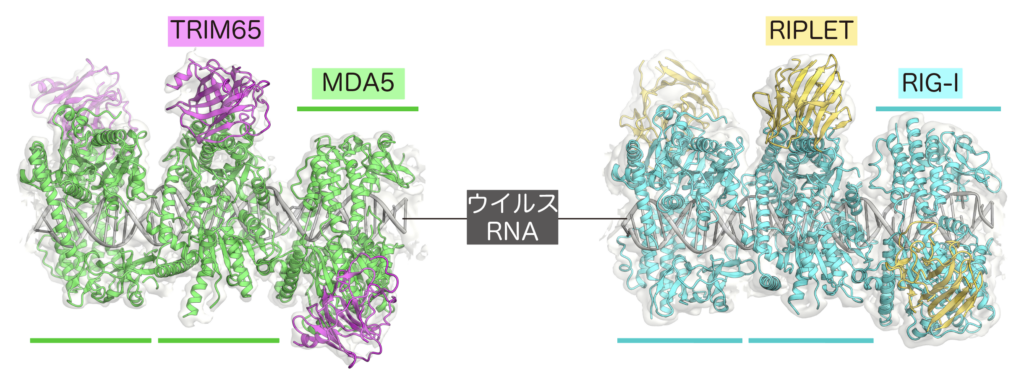
(左) MDA5-RNA-TRIM65.(右) RIG-I-RNA-RIPLET
2. Elucidation of the pathogenic mechanism of autoimmune diseases caused by RIG-I/MDA5 mutations and development of therapeutic agents
Mutations in RIG-I/MDA5 have been reported in humans. These RIG-I/MDA5 mutants, as gain-of-function mutants, misrecognize “self” RNA and elicit an immune response even in the absence of a viral infection. This results in autoimmune diseases such as Singleton-Merten syndrome, diabetes and Eckardt-Gutière syndrome. How do these RIG-I/MDA5 mutants misrecognize “self” RNA? Our research focuses on the mechanism. Based on the findings, we are developing therapeutic agents for autoimmune diseases.
3. Elucidation of self-antigen recognition mechanism by autoantibodies
Antibodies also directly recognize and eliminate bacteria and viruses as “non-self”. However, it has been reported that antibodies often misrecognize autoantigens in patients with autoimmune diseases. In our laboratory, we are actually isolating autoantibodies to elucidate the mechanism of misrecognition of autoantigens.
Development of novel tools utilizing prokaryotic antiphage defense mechanisms
Bacteria cause a variety of infectious diseases when they invade our bodies, but bacteria themselves are also threatened by phage viruses. It is thought that there are millions of species of prokaryotes (bacteria and archaea) on earth, each of which has evolved a variety of defense systems to counter phage virus infection. enzyme to suppress phage virus infection. A wide variety of Cas enzymes have been discovered and widely used from basic research in the life science field to the medical and industrial fields. For example, Cas9, which was discovered in the bacterium S. pyrogenes, cuts specific DNA sequences using guide RNA as a landmark, and Cas9 has caused a paradigm shift in the field of genome editing technology because it can freely rewrite genetic information in animals and plants as a “scissors” to cut genomic genes. Cas9 has caused a paradigm shift in the field of genome editing technology (Fig. 4).
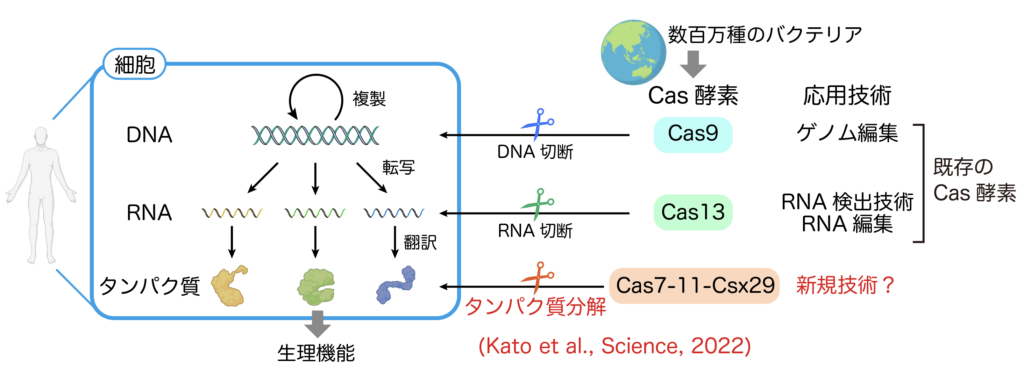
Cas enzymes discovered so far only cleave nucleic acids such as DNA and RNA, but no protein-cleaving Cas enzymes have been discovered. I discovered Cas7-11, a protease-type Cas enzyme, under the supervision of Professor Hiroshi Nishimasu of the Research Center for Advanced Science and Technology, the University of Tokyo (Kato et al., Science, 2022, https://www.science.org/doi/10.1126/ science.add7347). Cas7-11 forms a complex with Csx29 protease and guide RNA and shows protease activity when it recognizes target RNA (Figure 5). Through collaborative research with domestic and international academia and companies, our laboratory aims to apply the protease activity of Cas7-11-Csx29 to human cells to develop new technologies.
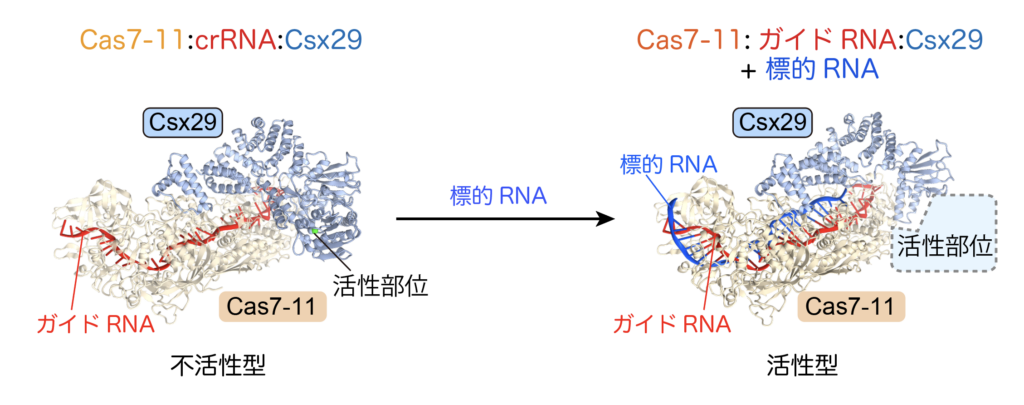
The Cas7-11-Csx29 complex normally exists as a protease-inactive form (left) and becomes active upon binding of the target RNA.
Research methods
In conducting these studies, we begin by expressing and purifying the protein under study as a recombinant protein.
(Biochemical analysis)
The purified proteins and RNAs are then mixed in vitro, and their functions are investigated in detail by biochemical experiments such as enzymatic reactions and binding experiments. In vitro, we can set reaction conditions such as the presence or absence of compounds, pH, and salt concentration as desired, allowing us to investigate the function of the target protein in detail. In this way, we search for new biological phenomena.
(Cell biological analysis)
Verification is performed by cell-based experiments under more physiological conditions. By gene transfer of the gene encoding the protein under study, either as a knockout or as a foreign plasmid, changes in the expression levels of downstream genes, immune responses and cell death are measured as outputs.
(Structural biological analysis)
By actually seeing the three-dimensional structure of the protein under study, we can learn more about its function and mechanism. In the past, X-ray crystallography was the main method for determining the three-dimensional structure of proteins at the atomic resolution level, but it sometimes took a lot of time and effort to crystallize the protein. Recently, it has become possible to easily determine the three-dimensional structure of proteins using cryo-electron microscopy. In our laboratory, we mainly use cryo-EM to determine the three-dimensional structures of novel biological phenomena discovered in biochemical and cellular experiments, and to investigate the detailed mechanisms of these phenomena.